Methods
Summary
Specific Aim 1: To identify highly conserved regions of the Zika virus genome to use for design of a diagnostic assay using quantitative reverse transcriptase PCR.
Experimental Approach: There are currently 44 complete Zika genome sequences in the GenBank database. We will align these publicly available Zika genomes against those of other related flaviviruses (like dengue) to identify regions of the genome that are conserved in all Zika isolates but not found in other flaviviruses, thus ensuring both the sensitivity and the specificity of our assay.
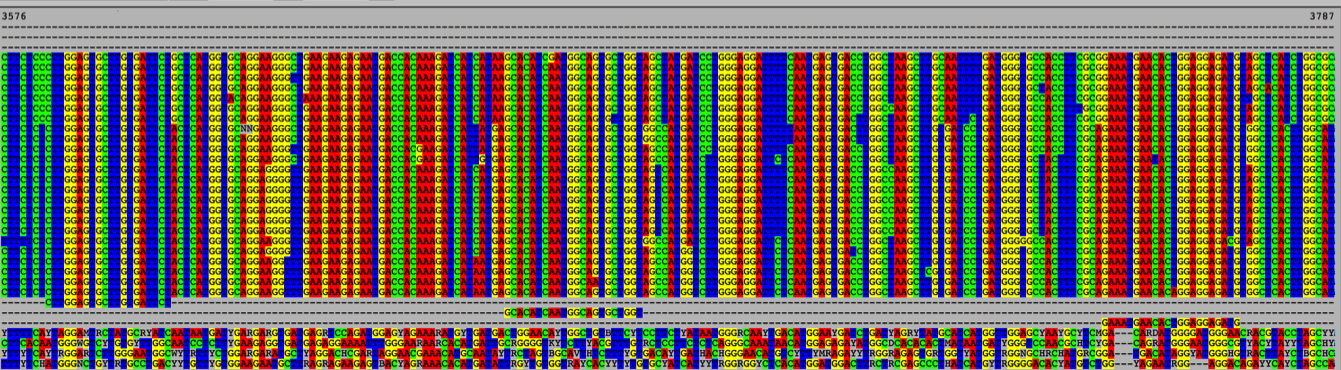 Figure 1. Example image of an alignment.
Figure 1. Example image of an alignment.
Specific Aim 2: Create and validate a quantitative reverse transcriptase PCR (qRT-PCR) capable of detecting the presence of even low concentrations of Zika RNA in mosquito samples.
Experimental Approach: Once the specific target sequences have been identified, we will deisgn specific primers to amplify this sequences and a probe to specifically to the sequence (TaqMan system). We will then run optimization plates to find the ideal primer, enzyme, and probe concentrations to amplify a broad range of template concentrations (hopefully down to the femtomolar range). To validate this assay we will then need to collect mosquitoes from the field, isolate total RNA, and use our newly developed assay to test for presence of Zika virus.
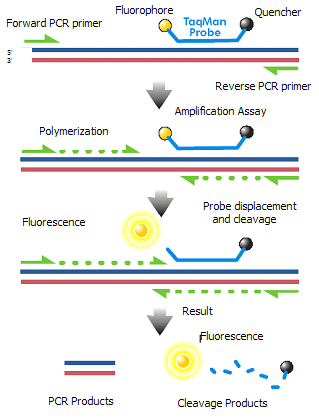
Figure 2. Schematic representation of qRT-PCR.
Specific Aim 3: Collect mosquito samples from Zika virus-endemic areas.
Experimental Approach: We aim to travel to areas endemic for the Zika virus areas (Haiti and South America) to deploy tested mosquito traps to collect mosquitoes from these areas. The trap used will likely be a CDC miniature light trap, which is specially designed for capturing Aedes mosquitoes- the vector of Zika virus.

Figure 3. Example of a CDC miniature light trap. Source
Protocols
This project has not yet shared any protocols.