74
0
0
Like?
Please wait...
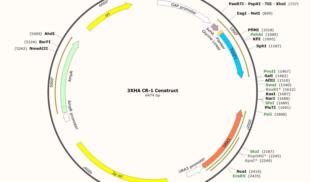
About This Project
Our project aims to provide a method of detecting pancreatic cancer in humans. We are using yeast in this project as a method for expressing Cripto-1, a membrane protein which binds to a biomarker for pancreatic cancer. We hope to be able to couple CR-1 binding to a reporter inside yeast for indication of the presence of the biomarker in human blood serum. This could offer insight into the potential for synthetic biology as an important medical tool in the future.

Browse Other Projects on Experiment
Related Projects
Disrupting cancer cell signaling through drug discovery
Most cancer-related deaths are caused by metastasis, the spread of cancer cells to distant tissues. This...
CaniSense– AI-powered blood test for early cancer detection in dogs
Cancer is the leading cause of death in dogs, yet no reliable methods for early screening exist. At testblu...
Shutting down cancer’s recycling system with exosome-based therapy
Pancreatic cancer is one of the deadliest cancers because its cells survive by recycling their own components...